AUTHORS:
This module defines a class of abstract finite cell complexes. This is meant as a base class from which other classes (like SimplicialComplex, CubicalComplex, and DeltaComplex) should derive. As such, most of its properties are not implemented. It is meant for use by developers producing new classes, not casual users.
Note
Keywords for GenericCellComplex.chain_complex(), GenericCellComplex.homology(), etc.: any keywords given to the homology() method get passed on to the chain_complex() method and also to the constructor for chain complexes in ChainComplex, as well its associated homology() method. This means that those keywords should have consistent meaning in all of those situations. It also means that it is easy to implement new keywords: for example, if you implement a new keyword for the sage.homology.chain_complex.ChainComplex.homology() method, then it will be automatically accessible through the homology() method for cell complexes – just make sure it gets documented.
Bases: sage.structure.sage_object.SageObject
Class of abstract cell complexes.
This is meant to be used by developers to produce new classes, not by casual users. Classes which derive from this are SimplicialComplex, DeltaComplex, and CubicalComplex.
Most of the methods here are not implemented, but probably should
be implemented in a derived class. Most of the other methods call
a non-implemented one; their docstrings contain examples from
derived classes in which the various methods have been defined.
For example, homology() calls chain_complex(); the
class DeltaComplex
implements
chain_complex(),
and so the homology() method here is illustrated with
examples involving  -complexes.
-complexes.
EXAMPLES:
It’s hard to give informative examples of the base class, since essentially nothing is implemented.
sage: from sage.homology.cell_complex import GenericCellComplex
sage: A = GenericCellComplex()
The Betti numbers of this simplicial complex as a dictionary (or a single Betti number, if only one dimension is given): the ith Betti number is the rank of the ith homology group.
| Parameters: |
|
|---|
EXAMPLES: Build the two-sphere as a three-fold join of a two-point space with itself:
sage: S = SimplicialComplex(1, [[0], [1]])
sage: (S*S*S).betti()
{0: 1, 1: 0, 2: 1}
sage: (S*S*S).betti([1,2])
{1: 0, 2: 1}
sage: (S*S*S).betti(2)
1
Or build the two-sphere as a  -complex:
-complex:
sage: S2 = delta_complexes.Sphere(2)
sage: S2.betti([1,2])
{1: 0, 2: 1}
Or as a cubical complex:
sage: S2c = cubical_complexes.Sphere(2)
sage: S2c.betti(2)
1
Return the category to which this chain complex belongs: the category of all cell complexes.
This is not implemented for general cell complexes.
EXAMPLES:
sage: from sage.homology.cell_complex import GenericCellComplex
sage: A = GenericCellComplex()
sage: A.category()
...
NotImplementedError
The cells of this cell complex, in the form of a dictionary: the keys are integers, representing dimension, and the value associated to an integer d is the set of d-cells. If the optional argument subcomplex is present, then return only the faces which are not in the subcomplex.
| Parameter: | subcomplex (optional, default None) – a subcomplex of this cell complex. Return the cells which are not in this subcomplex. |
|---|
This is not implemented in general; it should be implemented in any derived class. When implementing, see the warning in the dimension() method.
This method is used by various other methods, such as n_cells() and f_vector().
EXAMPLES:
sage: from sage.homology.cell_complex import GenericCellComplex
sage: A = GenericCellComplex()
sage: A.cells()
...
NotImplementedError
This is not implemented for general cell complexes.
Some keywords to possibly implement in a derived class:
Definitely implement the following:
EXAMPLES:
sage: from sage.homology.cell_complex import GenericCellComplex
sage: A = GenericCellComplex()
sage: A.chain_complex()
...
NotImplementedError
The reduced cohomology of this cell complex.
The arguments are the same as for the homology() method, except that homology() accepts a cohomology key word, while this function does not: cohomology is automatically true here. Indeed, this function just calls homology() with cohomology set to True.
| Parameters: |
|
|---|
EXAMPLES:
sage: circle = SimplicialComplex(2, [[0,1], [1,2], [0, 2]])
sage: circle.cohomology(0)
0
sage: circle.cohomology(1)
Z
sage: P2 = SimplicialComplex(5, [[0,1,2], [0,2,3], [0,1,5], [0,4,5], [0,3,4], [1,2,4], [1,3,4], [1,3,5], [2,3,5], [2,4,5]]) # projective plane
sage: P2.cohomology(2)
C2
sage: P2.cohomology(2, base_ring=GF(2))
Vector space of dimension 1 over Finite Field of size 2
sage: P2.cohomology(2, base_ring=GF(3))
Vector space of dimension 0 over Finite Field of size 3
sage: cubical_complexes.KleinBottle().cohomology(2)
C2
Relative cohomology:
sage: T = SimplicialComplex(1, [[0,1]])
sage: U = SimplicialComplex(1, [[0], [1]])
sage: T.cohomology(1, subcomplex=U)
Z
A  -complex example:
-complex example:
sage: s5 = delta_complexes.Sphere(5)
sage: s5.cohomology(base_ring=GF(7))[5]
Vector space of dimension 1 over Finite Field of size 7
The dimension of this cell complex: the maximum dimension of its cells.
Warning
If the cells() method calls dimension(), then you’ll get an infinite loop. So either don’t use dimension() or override dimension().
EXAMPLES:
sage: simplicial_complexes.RandomComplex(d=5, n=8).dimension()
5
sage: delta_complexes.Sphere(3).dimension()
3
sage: T = cubical_complexes.Torus()
sage: T.product(T).dimension()
4
The disjoint union of this simplicial complex with another one.
| Parameter: | right – the other simplicial complex (the right-hand factor) |
|---|
Disjoint unions are not implemented for general cell complexes.
EXAMPLES:
sage: from sage.homology.cell_complex import GenericCellComplex
sage: A = GenericCellComplex(); B = GenericCellComplex()
sage: A.disjoint_union(B)
...
NotImplementedError
The Euler characteristic of this cell complex: the
alternating sum over 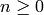 of the number of
of the number of
 -cells.
-cells.
EXAMPLES:
sage: simplicial_complexes.Simplex(5).euler_characteristic()
1
sage: delta_complexes.Sphere(6).euler_characteristic()
2
sage: cubical_complexes.KleinBottle().euler_characteristic()
0
The 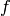 -vector of this cell complex: a list whose
-vector of this cell complex: a list whose 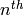 item is the number of
item is the number of 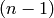 -cells. Note that, like all
lists in Sage, this is indexed starting at 0: the 0th element
in this list is the number of
-cells. Note that, like all
lists in Sage, this is indexed starting at 0: the 0th element
in this list is the number of 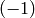 -cells (which is 1: the
empty cell is the only
-cells (which is 1: the
empty cell is the only 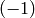 -cell).
-cell).
EXAMPLES:
sage: simplicial_complexes.KleinBottle().f_vector()
[1, 9, 27, 18]
sage: delta_complexes.KleinBottle().f_vector()
[1, 1, 3, 2]
sage: cubical_complexes.KleinBottle().f_vector()
[1, 42, 84, 42]
The face poset of this cell complex, the poset of nonempty cells, ordered by inclusion.
This uses the cells() method, and also assumes that for
each cell f, all of f.faces(), tuple(f), and
f.dimension() make sense. (If this is not the case in
some derived class, as happens with  -complexes, then
override this method.)
-complexes, then
override this method.)
EXAMPLES:
sage: P = SimplicialComplex(3, [[0, 1], [1,2], [2,3]]).face_poset(); P
Finite poset containing 7 elements
sage: P.list()
[(3,), (2,), (2, 3), (1,), (0,), (1, 2), (0, 1)]
sage: S2 = cubical_complexes.Sphere(2)
sage: S2.face_poset()
Finite poset containing 26 elements
The 1-skeleton of this cell complex, as a graph.
This is not implemented for general cell complexes.
EXAMPLES:
sage: from sage.homology.cell_complex import GenericCellComplex
sage: A = GenericCellComplex()
sage: A.graph()
...
NotImplementedError
The reduced homology of this cell complex.
| Parameters: |
|
|---|
Note
The keyword arguments to this function get passed on to :meth:chain_complex and its homology.
ALGORITHM:
If algorithm is set to ‘auto’ (the default), then use CHomP if available. (CHomP is available at the web page http://chomp.rutgers.edu/. It is also an experimental package for Sage.)
CHomP computes homology, not cohomology, and only works over the integers or finite prime fields. Therefore if any of these conditions fails, or if CHomP is not present, or if algorithm is set to ‘no_chomp’, go to plan B: if self has a _homology method – each simplicial complex has this, for example – then call that. Such a method implements specialized algorithms for the particular type of cell complex.
Otherwise, move on to plan C: compute the chain complex of self and compute its homology groups. To do this: over a field, just compute ranks and nullities, thus obtaining dimensions of the homology groups as vector spaces. Over the integers, compute Smith normal form of the boundary matrices defining the chain complex according to the value of algorithm. If algorithm is ‘auto’ or ‘no_chomp’, then for each relatively small matrix, use the standard Sage method, which calls the Pari package. For any large matrix, reduce it using the Dumas, Heckenbach, Saunders, and Welker elimination algorithm: see sage.homology.chain_complex.dhsw_snf() for details.
Finally, algorithm may also be ‘pari’ or ‘dhsw’, which forces the named algorithm to be used regardless of the size of the matrices and regardless of whether CHomP is available.
As of this writing, CHomP is by far the fastest option, followed by the ‘auto’ or ‘no_chomp’ setting of using the Dumas, Heckenbach, Saunders, and Welker elimination algorithm for large matrices and Pari for small ones.
EXAMPLES:
sage: P = delta_complexes.RealProjectivePlane()
sage: P.homology()
{0: 0, 1: C2, 2: 0}
sage: P.homology(base_ring=GF(2))
{0: Vector space of dimension 0 over Finite Field of size 2,
1: Vector space of dimension 1 over Finite Field of size 2,
2: Vector space of dimension 1 over Finite Field of size 2}
sage: S7 = delta_complexes.Sphere(7)
sage: S7.homology(7)
Z
sage: cubical_complexes.KleinBottle().homology(1, base_ring=GF(2))
Vector space of dimension 2 over Finite Field of size 2
If CHomP is installed, Sage can compute generators of homology groups:
sage: S2 = simplicial_complexes.Sphere(2)
sage: S2.homology(dim=2, generators=True) # optional: need CHomP
(Z, [(0, 1, 2) - (0, 1, 3) + (0, 2, 3) - (1, 2, 3)])
When generators are computed, Sage returns a pair for each dimension: the group and the list of generators. For simplicial complexes, each generator is represented as a linear combination of simplices, as above, and for cubical complexes, each generator is a linear combination of cubes:
sage: S2_cub = cubical_complexes.Sphere(2)
sage: S2_cub.homology(dim=2, generators=True) # optional: need CHomP
(Z, [-[[0,1] x [0,1] x [0,0]] + [[0,1] x [0,1] x [1,1]] - [[0,0] x [0,1] x [0,1]] - [[0,1] x [1,1] x [0,1]] + [[0,1] x [0,0] x [0,1]] + [[1,1] x [0,1] x [0,1]]])
The join of this cell complex with another one.
| Parameter: | right – the other simplicial complex (the right-hand factor) |
|---|
Joins are not implemented for general cell complexes. They may be implemented in some derived classes (like simplicial complexes).
EXAMPLES:
sage: from sage.homology.cell_complex import GenericCellComplex
sage: A = GenericCellComplex(); B = GenericCellComplex()
sage: A.join(B)
...
NotImplementedError
List of cells of dimension n of this cell complex. If the optional argument subcomplex is present, then return the n-dimensional faces which are not in the subcomplex.
| Parameters: |
|
|---|
EXAMPLES:
sage: simplicial_complexes.Simplex(2).n_cells(1)
[(1, 2), (0, 2), (0, 1)]
sage: delta_complexes.Torus().n_cells(1)
[(0, 0), (0, 0), (0, 0)]
sage: cubical_complexes.Cube(1).n_cells(0)
[[1,1], [0,0]]
The  -skeleton of this cell complex: the cell
complex obtained by discarding all of the simplices in
dimensions larger than
-skeleton of this cell complex: the cell
complex obtained by discarding all of the simplices in
dimensions larger than  .
.
| Parameter: | n – non-negative integer |
|---|
This is not implemented for general cell complexes.
EXAMPLES:
sage: from sage.homology.cell_complex import GenericCellComplex
sage: A = GenericCellComplex()
sage: A.n_skeleton(3)
...
NotImplementedError
The (Cartesian) product of this cell complex with another one.
Products are not implemented for general cell complexes. They may be implemented in some derived classes (like simplicial complexes).
EXAMPLES:
sage: from sage.homology.cell_complex import GenericCellComplex
sage: A = GenericCellComplex(); B = GenericCellComplex()
sage: A.product(B)
...
NotImplementedError
The wedge (one-point union) of this simplicial complex with another one.
| Parameter: | right – the other simplicial complex (the right-hand factor) |
|---|
Wedges are not implemented for general cell complexes.
EXAMPLES:
sage: from sage.homology.cell_complex import GenericCellComplex
sage: A = GenericCellComplex(); B = GenericCellComplex()
sage: A.wedge(B)
...
NotImplementedError