AUTHORS:
This module implements the basic structure of finite
 -complexes. For full mathematical details, see Hatcher [Hat],
especially Section 2.1 and the Appendix on “Simplicial CW Structures”.
As Hatcher points out,
-complexes. For full mathematical details, see Hatcher [Hat],
especially Section 2.1 and the Appendix on “Simplicial CW Structures”.
As Hatcher points out,  -complexes were first introduced by Eilenberg
and Zilber [EZ], although they called them “semi-simplicial complexes”.
-complexes were first introduced by Eilenberg
and Zilber [EZ], although they called them “semi-simplicial complexes”.
A  -complex is a generalization of a simplicial complex; a
-complex is a generalization of a simplicial complex; a  -complex
-complex 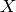 consists
of sets
consists
of sets 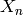 for each non-negative integer
for each non-negative integer  , the elements of which
are called n-simplices, along with face maps between these sets of
simplices: for each
, the elements of which
are called n-simplices, along with face maps between these sets of
simplices: for each  and for all
and for all 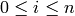 , there are
functions
, there are
functions 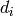 from
from 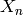 to
to 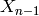 , with
, with 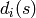 equal to the
equal to the
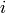 -th face of
-th face of  for each simplex
for each simplex 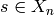 . These maps must
satisfy the simplicial identity
. These maps must
satisfy the simplicial identity
Given a  -complex, it has a geometric realization: a
topological space built by taking one topological
-complex, it has a geometric realization: a
topological space built by taking one topological  -simplex for each
element of
-simplex for each
element of 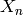 , and gluing them together as determined by the face
maps.
, and gluing them together as determined by the face
maps.
 -complexes are an alternative to simplicial complexes. Every
simplicial complex is automatically a
-complexes are an alternative to simplicial complexes. Every
simplicial complex is automatically a  -complex; in the other
direction, though, it seems in practice that one can often construct
-complex; in the other
direction, though, it seems in practice that one can often construct
 -complex representations for spaces with many fewer simplices
than in a simplicial complex representation. For example, the minimal
triangulation of a torus as a simplicial complex contains 14
triangles, 21 edges, and 7 vertices, while there is a
-complex representations for spaces with many fewer simplices
than in a simplicial complex representation. For example, the minimal
triangulation of a torus as a simplicial complex contains 14
triangles, 21 edges, and 7 vertices, while there is a  -complex
representation of a torus using only 2 triangles, 3 edges, and 1
vertex.
-complex
representation of a torus using only 2 triangles, 3 edges, and 1
vertex.
Note
This class derives from GenericCellComplex, and so inherits its methods. Some of those methods are not listed here; see the Generic Cell Complex page instead.
REFERENCES:
| [Hat] | Allen Hatcher, “Algebraic Topology”, Cambridge University Press (2002). |
| [EZ] | S. Eilenberg and J. Zilber, “Semi-Simplicial Complexes and Singular Homology”, Ann. Math. (2) 51 (1950), 499-513. |
Bases: sage.homology.cell_complex.GenericCellComplex
Define a  -complex.
-complex.
| Parameters: |
|
|---|---|
| Returns: | a |
Use data to define a  -complex. It may be in any of
three forms:
-complex. It may be in any of
three forms:
data may be a dictionary indexed by simplices. The value
associated to a d-simplex  can be any of:
can be any of:
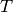 , in which case the ith face of
, in which case the ith face of  is
declared to be the same as the ith face of
is
declared to be the same as the ith face of 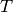 :
:  and
and  are glued along their entire boundary,
are glued along their entire boundary, .
.For example, consider the following:
sage: n = 5
sage: S5 = DeltaComplex({Simplex(n):True, Simplex(range(1,n+2)): Simplex(n)})
sage: S5
Delta complex with 6 vertices and 65 simplices
The first entry in dictionary forming the argument to
DeltaComplex says that there is an  -dimensional simplex
with its ordinary boundary. The second entry says that there is
another simplex whose boundary is glued to that of the first
one. The resulting
-dimensional simplex
with its ordinary boundary. The second entry says that there is
another simplex whose boundary is glued to that of the first
one. The resulting  -complex is, of course, homeomorphic
to an
-complex is, of course, homeomorphic
to an  -sphere, or actually a 5-sphere, since we defined
-sphere, or actually a 5-sphere, since we defined  to be 5. (Note that the second simplex here can be any
to be 5. (Note that the second simplex here can be any
 -dimensional simplex, as long as it is distinct from
Simplex(n).)
-dimensional simplex, as long as it is distinct from
Simplex(n).)
Let’s compute its homology, and also compare it to the simplicial version:
sage: S5.homology()
{0: 0, 1: 0, 2: 0, 3: 0, 4: 0, 5: Z}
sage: S5.f_vector() # number of simplices in each dimension
[1, 6, 15, 20, 15, 6, 2]
sage: simplicial_complexes.Sphere(5).f_vector()
[1, 7, 21, 35, 35, 21, 7]
Both contain a single (-1)-simplex, the empty simplex; other
than that, the  -complex version contains fewer simplices
than the simplicial one in each dimension.
-complex version contains fewer simplices
than the simplicial one in each dimension.
To construct a torus, use:
sage: torus_dict = {Simplex([0,1,2]): True,
... Simplex([3,4,5]): (Simplex([0,1]), Simplex([0,2]), Simplex([1,2])),
... Simplex([0,1]): (Simplex(0), Simplex(0)),
... Simplex([0,2]): (Simplex(0), Simplex(0)),
... Simplex([1,2]): (Simplex(0), Simplex(0)),
... Simplex(0): ()}
sage: T = DeltaComplex(torus_dict); T
Delta complex with 1 vertex and 7 simplices
sage: T.cohomology(base_ring=QQ)
{0: Vector space of dimension 0 over Rational Field,
1: Vector space of dimension 2 over Rational Field,
2: Vector space of dimension 1 over Rational Field}
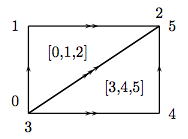
This  -complex consists of two triangles (given by
Simplex([0,1,2]) and Simplex([3,4,5])); the boundary of
the first is just its usual boundary: the 0th face is obtained
by omitting the lowest numbered vertex, etc., and so the
boundary consists of the edges [1,2], [0,2], and
[0,1], in that order. The boundary of the second is, on the
one hand, computed the same way: the nth face is obtained by
omitting the nth vertex. On the other hand, the boundary is
explicitly declared to be edges [0,1], [0,2], and
[1,2], in that order. This glues the second triangle to the
first in the prescribed way. The three edges each start and end
at the single vertex, Simplex(0).
-complex consists of two triangles (given by
Simplex([0,1,2]) and Simplex([3,4,5])); the boundary of
the first is just its usual boundary: the 0th face is obtained
by omitting the lowest numbered vertex, etc., and so the
boundary consists of the edges [1,2], [0,2], and
[0,1], in that order. The boundary of the second is, on the
one hand, computed the same way: the nth face is obtained by
omitting the nth vertex. On the other hand, the boundary is
explicitly declared to be edges [0,1], [0,2], and
[1,2], in that order. This glues the second triangle to the
first in the prescribed way. The three edges each start and end
at the single vertex, Simplex(0).
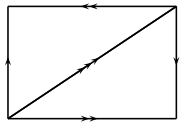
data may be nested lists or tuples. The nth entry in the list is a list of the n-simplices in the complex, and each n-simplex is encoded as a list, the ith entry of which is its ith face. Each face is represented by an integer, giving its index in the list of (n-1)-faces. For example, consider this:
sage: P = DeltaComplex( [ [(), ()], [(1,0), (1,0), (0,0)],
... [(1,0,2), (0, 1, 2)] ])
The 0th entry in the list is [(), ()]: there are two 0-simplices, and their boundaries are empty.
The 1st entry in the list is [(1,0), (1,0), (0,0)]: there are three 1-simplices. Two of them have boundary (1,0), which means that their 0th face is vertex 1 (in the list of vertices), and their 1st face is vertex 0. The other edge has boundary (0,0), so it starts and ends at vertex 0.
The 2nd entry in the list is [(1,0,2), (0,1,2)]: there are two 2-simplices. The first 2-simplex has boundary (1,0,2), meaning that its 0th face is edge 1 (in the list above), its 1st face is edge 0, and its 2nd face is edge 2; similarly for the 2nd 2-simplex.
If one draws two triangles and identifies them according to this description, the result is the real projective plane.
sage: P.homology(1)
C2
sage: P.cohomology(2)
C2
Closely related to this form for data is X.cells()
for a  -complex X: this is a dictionary, indexed by
dimension d, whose d-th entry is a list of the
d-simplices, as a list:
-complex X: this is a dictionary, indexed by
dimension d, whose d-th entry is a list of the
d-simplices, as a list:
sage: P.cells()
{0: ((), ()), 1: ((1, 0), (1, 0), (0, 0)), 2: ((1, 0, 2), (0, 1, 2)), -1: ((),)}
data may be a dictionary indexed by integers. For each
integer  , the entry with key
, the entry with key  is the list of
is the list of
 -simplices: this is the same format as is output by the
cells() method.
-simplices: this is the same format as is output by the
cells() method.
sage: P = DeltaComplex( [ [(), ()], [(1,0), (1,0), (0,0)],
... [(1,0,2), (0, 1, 2)] ])
sage: cells_dict = P.cells()
sage: cells_dict
{0: ((), ()), 1: ((1, 0), (1, 0), (0, 0)), 2: ((1, 0, 2), (0, 1, 2)), -1: ((),)}
sage: DeltaComplex(cells_dict)
Delta complex with 2 vertices and 8 simplices
sage: P == DeltaComplex(cells_dict)
True
Since  -complexes are generalizations of simplicial
complexes, any simplicial complex may be viewed as a
-complexes are generalizations of simplicial
complexes, any simplicial complex may be viewed as a
 -complex:
-complex:
sage: RP2 = simplicial_complexes.RealProjectivePlane()
sage: RP2_delta = RP2.delta_complex()
sage: RP2.f_vector()
[1, 6, 15, 10]
sage: RP2_delta.f_vector()
[1, 6, 15, 10]
Finally,  -complex constructions for several familiar
spaces are available as follows:
-complex constructions for several familiar
spaces are available as follows:
sage: delta_complexes.Sphere(4) # the 4-sphere
Delta complex with 5 vertices and 33 simplices
sage: delta_complexes.KleinBottle()
Delta complex with 1 vertex and 7 simplices
sage: delta_complexes.RealProjectivePlane()
Delta complex with 2 vertices and 8 simplices
Type delta_complexes. and then hit the TAB key to get the full list.
Not implemented.
EXAMPLES:
sage: K = delta_complexes.KleinBottle()
sage: K.barycentric_subdivision()
...
NotImplementedError: Barycentric subdivisions are not implemented for Delta complexes.
The cells of this  -complex.
-complex.
| Parameter: | subcomplex (optional, default None) – a subcomplex of this complex |
|---|
The cells of this  -complex, in the form of a dictionary:
the keys are integers, representing dimension, and the value
associated to an integer d is the list of d-cells. Each
d-cell is further represented by a list, the ith entry of
which gives the index of its ith face in the list of
(d-1)-cells.
-complex, in the form of a dictionary:
the keys are integers, representing dimension, and the value
associated to an integer d is the list of d-cells. Each
d-cell is further represented by a list, the ith entry of
which gives the index of its ith face in the list of
(d-1)-cells.
If the optional argument subcomplex is present, then “return only the faces which are not in the subcomplex”. To preserve the indexing, which is necessary to compute the relative chain complex, this actually replaces the faces in subcomplex with None.
EXAMPLES:
sage: S2 = delta_complexes.Sphere(2)
sage: S2.cells()
{0: ((), (), ()), 1: ((0, 1), (0, 2), (1, 2)), 2: ((0, 1, 2), (0, 1, 2)), -1: ((),)}
sage: A = S2.subcomplex({1: [0,2]}) # one edge
sage: S2.cells(subcomplex=A)
{0: (None, None, None), 1: (None, (0, 2), None), 2: ((0, 1, 2), (0, 1, 2)), -1: (None,)}
The chain complex associated to this  -complex.
-complex.
| Parameters: |
|
|---|
Note
If subcomplex is nonempty, then the argument augmented
has no effect: the chain complex relative to a nonempty
subcomplex is zero in dimension 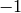 .
.
EXAMPLES:
sage: circle = delta_complexes.Sphere(1)
sage: circle.chain_complex()
Chain complex with at most 2 nonzero terms over Integer Ring
sage: circle.chain_complex()._latex_()
'\\Bold{Z}^{1} \\xrightarrow{d_{1}} \\Bold{Z}^{1}'
sage: circle.chain_complex(base_ring=QQ, augmented=True)
Chain complex with at most 3 nonzero terms over Rational Field
sage: circle.homology(dim=1)
Z
sage: circle.cohomology(dim=1)
Z
sage: T = T = delta_complexes.Torus()
sage: T.chain_complex(subcomplex=T)
Chain complex with at most 0 nonzero terms over Integer Ring
sage: T.homology(subcomplex=T)
{0: 0, 1: 0, 2: 0}
sage: A = T.subcomplex({2: [1]}) # one of the two triangles forming T
sage: T.chain_complex(subcomplex=A)
Chain complex with at most 1 nonzero terms over Integer Ring
sage: T.homology(subcomplex=A)
{0: 0, 1: 0, 2: Z}
The cone on this  -complex.
-complex.
The cone is the complex formed by adding a new vertex 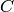 and
simplices of the form
and
simplices of the form ![[C, v_0, ..., v_k]](../../_images/math/965d465f2c17bbdc56454c318395dd6915107058.png) for every simplex
for every simplex
![[v_0, ..., v_k]](../../_images/math/667622c214dbe572cd4e9082e87ff5075cc8f0af.png) in the original complex. That is, the cone
is the join of the original complex with a one-point complex.
in the original complex. That is, the cone
is the join of the original complex with a one-point complex.
EXAMPLES:
sage: K = delta_complexes.KleinBottle()
sage: K.cone()
Delta complex with 2 vertices and 14 simplices
sage: K.cone().homology()
{0: 0, 1: 0, 2: 0, 3: 0}
Return the connected sum of self with other.
| Parameter: | other – another  -complex -complex |
|---|---|
| Returns: | the connected sum self # other |
Warning
This does not check that self and other are manifolds. It doesn’t even check that their facets all have the same dimension. It just chooses top-dimensional simplices from each complex, checks that they have the same dimension, removes them, and glues the remaining pieces together. Since a (more or less) random facet is chosen from each complex, this method may return random results if applied to non-manifolds, depending on which facet is chosen.
ALGORITHM:
Pick a top-dimensional simplex from each complex. Check to see if there are any identifications on either simplex, using the _is_glued() method. If there are no identifications, remove the simplices and glue the remaining parts of complexes along their boundary. If there are identifications on a simplex, subdivide it repeatedly (using elementary_subdivision()) until some piece has no identifications.
EXAMPLES:
sage: T = delta_complexes.Torus()
sage: S2 = delta_complexes.Sphere(2)
sage: T.connected_sum(S2).cohomology() == T.cohomology()
True
sage: RP2 = delta_complexes.RealProjectivePlane()
sage: T.connected_sum(RP2).homology(1)
Z x Z x C2
sage: T.connected_sum(RP2).homology(2)
0
sage: RP2.connected_sum(RP2).connected_sum(RP2).homology(1)
Z x Z x C2
The disjoint union of this  -complex with another one.
-complex with another one.
| Parameter: | right – the other  -complex (the right-hand factor) -complex (the right-hand factor) |
|---|
EXAMPLES:
sage: S1 = delta_complexes.Sphere(1)
sage: S2 = delta_complexes.Sphere(2)
sage: S1.disjoint_union(S2).homology()
{0: Z, 1: Z, 2: Z}
Perform an “elementary subdivision” on a top-dimensional
simplex in this  -complex. If the optional argument
idx is present, it specifies the index (in the list of
top-dimensional simplices) of the simplex to subdivide. If
not present, subdivide the last entry in this list.
-complex. If the optional argument
idx is present, it specifies the index (in the list of
top-dimensional simplices) of the simplex to subdivide. If
not present, subdivide the last entry in this list.
| Parameter: | idx (integer; optional, default -1) – index specifying which simplex to subdivide |
|---|---|
| Returns: |  -complex with one simplex subdivided. -complex with one simplex subdivided. |
Elementary subdivision of a simplex means replacing that
simplex with the cone on its boundary. That is, given a
 -complex containing an
-complex containing an 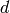 -simplex
-simplex  with vertices
with vertices
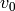 , ...,
, ..., 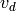 , form a new
, form a new  -complex by
-complex by

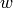 (thought of as being in the interior of
(thought of as being in the interior of  )
)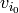 , ...,
, ...,
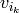 ,
, 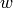 , preserving any identifications present
along the boundary of
, preserving any identifications present
along the boundary of 
The algorithm for achieving this uses
_epi_from_standard_simplex() to keep track of simplices
(with multiplicity) and what their faces are: this method
defines a surjection 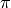 from the standard
from the standard 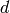 -simplex to
-simplex to
 . So first remove
. So first remove  and add a new vertex
and add a new vertex 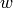 , say at the
end of the old list of vertices. Then for each vertex
, say at the
end of the old list of vertices. Then for each vertex  in
the standard
in
the standard 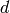 -simplex, add an edge from
-simplex, add an edge from 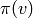 to
to 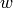 ;
for each edge
;
for each edge 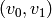 in the standard
in the standard 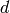 -simplex, add a
triangle
-simplex, add a
triangle 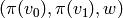 , etc.
, etc.
Note that given an  -simplex
-simplex 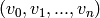 in the
standard
in the
standard 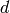 -simplex, the faces of the new
-simplex, the faces of the new 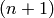 -simplex are
given by removing vertices, one at a time, from
-simplex are
given by removing vertices, one at a time, from 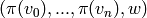 . These are either the image of the old
. These are either the image of the old
 -simplex (if
-simplex (if 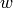 is removed) or the various new
is removed) or the various new
 -simplices added in the previous dimension. So keep track
of what’s added in dimension
-simplices added in the previous dimension. So keep track
of what’s added in dimension  for use in computing the
faces in dimension
for use in computing the
faces in dimension 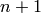 .
.
In contrast with barycentric subdivision, note that only the
interior of  has been changed; this allows for subdivision
of a single top-dimensional simplex without subdividing every
simplex in the complex.
has been changed; this allows for subdivision
of a single top-dimensional simplex without subdividing every
simplex in the complex.
The term “elementary subdivison” is taken from p. 112 in John M. Lee’s book [Lee].
REFERENCES:
| [Lee] | John M. Lee, Introduction to Topological Manifolds, Springer-Verlag, GTM volume 202. |
EXAMPLES:
sage: T = delta_complexes.Torus()
sage: T.n_cells(2)
[(1, 2, 0), (0, 2, 1)]
sage: T.elementary_subdivision(0) # subdivide first triangle
Delta complex with 2 vertices and 13 simplices
sage: X = T.elementary_subdivision(); X # subdivide last triangle
Delta complex with 2 vertices and 13 simplices
sage: X.elementary_subdivision()
Delta complex with 3 vertices and 19 simplices
sage: X.homology() == T.homology()
True
The face poset of this  -complex, the poset of
nonempty cells, ordered by inclusion.
-complex, the poset of
nonempty cells, ordered by inclusion.
EXAMPLES:
sage: T = delta_complexes.Torus()
sage: T.face_poset()
Finite poset containing 6 elements
The 1-skeleton of this  -complex as a graph.
-complex as a graph.
EXAMPLES:
sage: T = delta_complexes.Torus()
sage: T.graph()
Looped multi-graph on 1 vertex
sage: S = delta_complexes.Sphere(2)
sage: S.graph()
Graph on 3 vertices
sage: delta_complexes.Simplex(4).graph() == graphs.CompleteGraph(5)
True
The join of this  -complex with another one.
-complex with another one.
| Parameter: | other – another  -complex (the right-hand
factor) -complex (the right-hand
factor) |
|---|---|
| Returns: | the join self * other |
The join of two  -complexes
-complexes  and
and 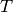 is the
is the
 -complex
-complex 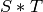 with simplices of the form
with simplices of the form ![[v_0, ...,
v_k, w_0, ..., w_n]](../../_images/math/fb1b043e0b46494e1c52071152ad23cdb735a591.png) for all simplices
for all simplices ![[v_0, ..., v_k]](../../_images/math/667622c214dbe572cd4e9082e87ff5075cc8f0af.png) in
in
 and
and ![[w_0, ..., w_n]](../../_images/math/f522ac0086a19ea0e89dbe2fbc57c846a80b67c6.png) in
in 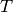 . The faces are computed
accordingly: the ith face of such a simplex is either
. The faces are computed
accordingly: the ith face of such a simplex is either 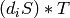 if
if 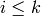 , or
, or 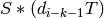 if
if 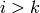 .
.
EXAMPLES:
sage: T = delta_complexes.Torus()
sage: S0 = delta_complexes.Sphere(0)
sage: T.join(S0) # the suspension of T
Delta complex with 3 vertices and 21 simplices
Compare to simplicial complexes:
sage: K = delta_complexes.KleinBottle()
sage: T_simp = simplicial_complexes.Torus()
sage: K_simp = simplicial_complexes.KleinBottle()
sage: T.join(K).homology()[3] == T_simp.join(K_simp).homology()[3] # long time (3 seconds)
True
The notation ‘*’ may be used, as well:
sage: S1 = delta_complexes.Sphere(1)
sage: X = S1 * S1 # X is a 3-sphere
sage: X.homology()
{0: 0, 1: 0, 2: 0, 3: Z}
The n-skeleton of this  -complex.
-complex.
| Parameter: | n (non-negative integer) – dimension |
|---|
EXAMPLES:
sage: S3 = delta_complexes.Sphere(3)
sage: S3.n_skeleton(1) # 1-skeleton of a tetrahedron
Delta complex with 4 vertices and 11 simplices
sage: S3.n_skeleton(1).dimension()
1
sage: S3.n_skeleton(1).homology()
{0: 0, 1: Z x Z x Z}
The product of this  -complex with another one.
-complex with another one.
| Parameter: | other – another  -complex (the right-hand
factor) -complex (the right-hand
factor) |
|---|---|
| Returns: | the product self x other |
Warning
If X and Y are  -complexes, then X*Y
returns their join, not their product.
-complexes, then X*Y
returns their join, not their product.
EXAMPLES:
sage: K = delta_complexes.KleinBottle()
sage: X = K.product(K)
sage: X.homology(1)
Z x Z x C2 x C2
sage: X.homology(2)
Z x C2 x C2 x C2
sage: X.homology(3)
C2
sage: X.homology(4)
0
sage: X.homology(base_ring=GF(2))
{0: Vector space of dimension 0 over Finite Field of size 2,
1: Vector space of dimension 4 over Finite Field of size 2,
2: Vector space of dimension 6 over Finite Field of size 2,
3: Vector space of dimension 4 over Finite Field of size 2,
4: Vector space of dimension 1 over Finite Field of size 2}
sage: S1 = delta_complexes.Sphere(1)
sage: K.product(S1).homology() == S1.product(K).homology()
True
sage: S1.product(S1) == delta_complexes.Torus()
True
Create a subcomplex.
| Parameter: | data – a dictionary indexed by dimension or a list (or tuple); in either case, data[n] should be the list (or tuple or set) of the indices of the simplices to be included in the subcomplex. |
|---|
This automatically includes all faces of the simplices in data, so you only have to specify the simplices which are maximal with respect to inclusion.
EXAMPLES:
sage: X = delta_complexes.Torus()
sage: A = X.subcomplex({2: [0]}) # one of the triangles of X
sage: X.homology(subcomplex=A)
{0: 0, 1: 0, 2: Z}
In the following, line is a line segment and ends is the complex consisting of its two endpoints, so the relative homology of the two is isomorphic to the homology of a circle:
sage: line = delta_complexes.Simplex(1) # an edge
sage: line.cells()
{0: ((), ()), 1: ((0, 1),), -1: ((),)}
sage: ends = line.subcomplex({0: (0, 1)})
sage: ends.cells()
{0: ((), ()), -1: ((),)}
sage: line.homology(subcomplex=ends)
{0: 0, 1: Z}
The suspension of this  -complex.
-complex.
| Parameter: | n (positive integer; optional, default 1) – suspend this many times. |
|---|
The suspension is the complex formed by adding two new
vertices 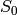 and
and 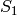 and simplices of the form
and simplices of the form ![[S_0, v_0,
..., v_k]](../../_images/math/8136f34cb4be566200a5c697f25886b39280c97f.png) and
and ![[S_1, v_0, ..., v_k]](../../_images/math/c13ce6f9dc3a1f6e60bcb457b93877aef7e6d0f8.png) for every simplex
for every simplex ![[v_0,
..., v_k]](../../_images/math/94c37dae2a62275a635a0f44f0a524e1e0472d01.png) in the original complex. That is, the suspension
is the join of the original complex with a two-point complex
(the 0-sphere).
in the original complex. That is, the suspension
is the join of the original complex with a two-point complex
(the 0-sphere).
EXAMPLES:
sage: S = delta_complexes.Sphere(0)
sage: S3 = S.suspension(3) # the 3-sphere
sage: S3.homology()
{0: 0, 1: 0, 2: 0, 3: Z}
The wedge (one-point union) of this  -complex with
another one.
-complex with
another one.
| Parameter: | right – the other  -complex (the right-hand factor) -complex (the right-hand factor) |
|---|
Note
This operation is not well-defined if self or other is not path-connected.
EXAMPLES:
sage: S1 = delta_complexes.Sphere(1)
sage: S2 = delta_complexes.Sphere(2)
sage: S1.wedge(S2).homology()
{0: 0, 1: Z, 2: Z}
Some examples of  -complexes.
-complexes.
Here are the available examples; you can also type delta_complexes. and hit TAB to get a list:
Sphere
Torus
RealProjectivePlane
KleinBottle
Simplex
SurfaceOfGenus
EXAMPLES:
sage: S = delta_complexes.Sphere(6) # the 6-sphere
sage: S.dimension()
6
sage: S.cohomology(6)
Z
sage: delta_complexes.Torus() == delta_complexes.Sphere(3)
False
A  -complex representation of the Klein bottle, consisting
of one vertex, three edges, and two triangles.
-complex representation of the Klein bottle, consisting
of one vertex, three edges, and two triangles.
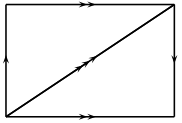
EXAMPLES:
sage: delta_complexes.KleinBottle()
Delta complex with 1 vertex and 7 simplices
A  -complex representation of the real projective plane,
consisting of two vertices, three edges, and two triangles.
-complex representation of the real projective plane,
consisting of two vertices, three edges, and two triangles.

EXAMPLES:
sage: P = delta_complexes.RealProjectivePlane()
sage: P.cohomology(1)
0
sage: P.cohomology(2)
C2
sage: P.cohomology(dim=1, base_ring=GF(2))
Vector space of dimension 1 over Finite Field of size 2
sage: P.cohomology(dim=2, base_ring=GF(2))
Vector space of dimension 1 over Finite Field of size 2
A  -complex representation of an
-complex representation of an  -simplex,
consisting of a single
-simplex,
consisting of a single  -simplex and its faces. (This is
the same as the simplicial complex representation available by
using simplicial_complexes.Simplex(n).)
-simplex and its faces. (This is
the same as the simplicial complex representation available by
using simplicial_complexes.Simplex(n).)
EXAMPLES:
sage: delta_complexes.Simplex(3)
Delta complex with 4 vertices and 16 simplices
A  -complex representation of the
-complex representation of the  -dimensional sphere,
formed by gluing two
-dimensional sphere,
formed by gluing two  -simplices along their boundary,
except in dimension 1, in which case it is a single 1-simplex
starting and ending at the same vertex.
-simplices along their boundary,
except in dimension 1, in which case it is a single 1-simplex
starting and ending at the same vertex.
| Parameter: | n – dimension of the sphere |
|---|
EXAMPLES:
sage: delta_complexes.Sphere(4).cohomology(4, base_ring=GF(3))
Vector space of dimension 1 over Finite Field of size 3
A surface of genus g as a  -complex.
-complex.
| Parameters: |
|
|---|
In the orientable case, return a sphere if 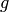 is zero, and
otherwise return a
is zero, and
otherwise return a 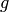 -fold connected sum of a torus with
itself.
-fold connected sum of a torus with
itself.
In the non-orientable case, raise an error if 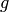 is zero. If
is zero. If
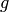 is positive, return a
is positive, return a 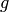 -fold connected sum of a
real projective plane with itself.
-fold connected sum of a
real projective plane with itself.
EXAMPLES:
sage: delta_complexes.SurfaceOfGenus(1, orientable=False)
Delta complex with 2 vertices and 8 simplices
sage: delta_complexes.SurfaceOfGenus(3, orientable=False).homology(1)
Z x Z x C2
sage: delta_complexes.SurfaceOfGenus(3, orientable=False).homology(2)
0
Compare to simplicial complexes:
sage: delta_g4 = delta_complexes.SurfaceOfGenus(4)
sage: delta_g4.f_vector()
[1, 5, 33, 22]
sage: simpl_g4 = simplicial_complexes.SurfaceOfGenus(4)
sage: simpl_g4.f_vector()
[1, 19, 75, 50]
sage: delta_g4.homology() == simpl_g4.homology()
True
A  -complex representation of the torus, consisting of one
vertex, three edges, and two triangles.
-complex representation of the torus, consisting of one
vertex, three edges, and two triangles.
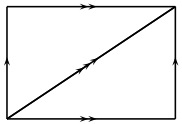
EXAMPLES:
sage: delta_complexes.Torus().homology(1)
Z x Z