See the Sage wiki page http://wiki.sagemath.org/graph_survey for an excellent survey of exisiting graph theory software.
Networkx (http://networkx.lanl.gov) “is a Python package for the creation, manipulation, and study of the structure, dynamics, and functions of complex networks”. More details can also be found on http://wiki.sagemath.org/graph_survey or in Robert Miller’s SageDays 3 talk.
sage: C = graphs.CubeGraph(4)
Now type C.show(vertex_labels=False, node_size=60, graph_border=True, figsize=[9,8]) to view this with some of the options.
The digraph below is a 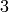 -cycle with vertices
-cycle with vertices
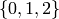 and edges
and edges 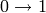 ,
,
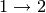 ,
, 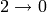 :
:
sage: D = DiGraph( { 0: [1], 1: [2], 2: [0]} )
Type D.show() to view this.
includes wrappers to many NetworkX commands, written mainly by Emily Kirkman and Robert Miller. The implementation of Cayley graphs was written by Bobby Moretti and Robert Miller.
sage: G = DihedralGroup(5)
sage: C = G.cayley_graph(); C
Digraph on 10 vertices
sage: C.diameter()
3
sage: C.girth()
4
sage: C.automorphism_group().order()
10
sage: len(C.edges())
20
To construct the graph G with 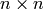 adjacency
matrix
adjacency
matrix 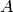 , you want a graph
, you want a graph 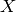 so that the
vertex-set of G is
so that the
vertex-set of G is 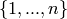 , and
, and ![[i,j]](_images/math/9467be6543ae7dfc59f1555f3f00952e4f3a00e6.png) is an edge of G if and only if
is an edge of G if and only if ![A[i][j] = 1](_images/math/81fd7de5e55f845fecf96f3690400a4e225a66a9.png) .
.
Here is an example of the syntax in (copied from Robert Miller’s
SageDays 3 talk): Define the distance 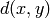 from
from 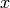 to
to
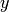 to be the minimum length of a (directed) path in Gamma
joining a vertex
to be the minimum length of a (directed) path in Gamma
joining a vertex 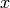 to a vertex
to a vertex 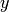 if such a path
exists, and
if such a path
exists, and 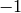 otherwise.
A diameter of
otherwise.
A diameter of 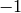 is returned if G is not (strongly)
connected. Otherwise, the diameter of G is equal to the maximum
(directed) distance
is returned if G is not (strongly)
connected. Otherwise, the diameter of G is equal to the maximum
(directed) distance 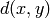 in G (as
in G (as 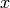 and
and
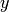 range over all the vertices of G).
range over all the vertices of G).
sage: M = Matrix ([ [0, 1, 1], [1, 0, 1], [1, 1, 0] ])
sage: # (the order is the number of edges)
sage: G = Graph(M); G.order()
3
sage: G.distance(0,2)
1
sage: G.diameter()
1