AUTHORS:
Bases: object
ode_solver() is a class that wraps the GSL libraries ode solver routines To use it instantiate a class,:
sage: T=ode_solver()
To solve a system of the form dy_i/dt=f_i(t,y), you must
supply a vector or tuple/list valued function f representing
f_i. The functions f and the jacobian should have the
form foo(t,y) or foo(t,y,params). params which is
optional allows for your function to depend on one or a tuple of
parameters. Note if you use it, params must be a tuple even
if it only has one component. For example if you wanted to solve
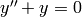 . You need to write it as a first order system:
. You need to write it as a first order system:
y_0' = y_1
y_1' = -y_0
In code:
sage: f = lambda t,y:[y[1],-y[0]]
sage: T.function=f
For some algorithms the jacobian must be supplied as well, the form of this should be a function return a list of lists of the form [ [df_1/dy_1,...,df_1/dy_n], ..., [df_n/dy_1,...,df_n,dy_n], [df_1/dt,...,df_n/dt] ].
There are examples below, if your jacobian was the function my_jacobian you would do:
sage: T.jacobian = my_jacobian # not tested, since it doesn't make sense to test this
There are a variety of algorithms available for different types of systems. Possible algorithms are
The default algorithm is rkf45. If you instead wanted to use bsimp you would do:
sage: T.algorithm="bsimp"
The user should supply initial conditions in y_0. For example if your initial conditions are y_0=1,y_1=1, do:
sage: T.y_0=[1,1]
The actual solver is invoked by the method ode_solve(). It has arguments t_span, y_0, num_points, params. y_0 must be supplied either as an argument or above by assignment. Params which are optional and only necessary if your system uses params can be supplied to ode_solve or by assignment.
t_span is the time interval on which to solve the ode. If t_span is a tuple with just two time values then the user must specify num_points, and the system will be evaluated at num_points equally spaced points between t_span[0] and t_span[1]. If t_span is a tuple with more than two values than the values of y_i at points in time listed in t_span will be returned.
Error is estimated via the expression D_i = error_abs*s_i+error_rel*(a|y_i|+a_dydt*h*|y_i'|). The user can specify error_abs (1e-10 by default), error_rel (1e-10 by default) a (1 by default), a_(dydt) (0 by default) and s_i (as scaling_abs which should be a tuple and is 1 in all components by default). If you specify one of a or a_dydt you must specify the other. You may specify a and a_dydt without scaling_abs (which will be taken =1 be default). h is the initial step size which is (1e-2) by default.
ode_solve solves the solution as a list of tuples of the form, [ (t_0,[y_1,...,y_n]),(t_1,[y_1,...,y_n]),...,(t_n,[y_1,...,y_n])].
This data is stored in the variable solutions:
sage: T.solution # not tested
EXAMPLES:
Consider solving the Van der Pol oscillator 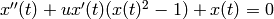 between
between 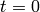 and
and 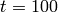 . As a first
order system it is
. As a first
order system it is 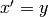 ,
, 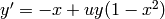 . Let us take
. Let us take 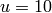 and use initial conditions
and use initial conditions 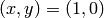 and use the runga-kutta
prince-dormand algorithm.
and use the runga-kutta
prince-dormand algorithm.
sage: def f_1(t,y,params):
... return[y[1],-y[0]-params[0]*y[1]*(y[0]**2-1.0)]
sage: def j_1(t,y,params):
... return [ [0.0, 1.0],[-2.0*params[0]*y[0]*y[1]-1.0,-params[0]*(y[0]*y[0]-1.0)] ]
sage: T=ode_solver()
sage: T.algorithm="rk8pd"
sage: T.function=f_1
sage: T.jacobian=j_1
sage: T.ode_solve(y_0=[1,0],t_span=[0,100],params=[10.0],num_points=1000)
sage: outfile = SAGE_TMP + 'sage.png'
sage: T.plot_solution(filename=outfile)
The solver line is equivalent to:
sage: T.ode_solve(y_0=[1,0],t_span=[x/10.0 for x in range(1000)],params = [10.0])
Let’s try a system:
y_0'=y_1*y_2
y_1'=-y_0*y_2
y_2'=-.51*y_0*y_1
We will not use the jacobian this time and will change the error tolerances.
sage: g_1= lambda t,y: [y[1]*y[2],-y[0]*y[2],-0.51*y[0]*y[1]]
sage: T.function=g_1
sage: T.y_0=[0,1,1]
sage: T.scale_abs=[1e-4,1e-4,1e-5]
sage: T.error_rel=1e-4
sage: T.ode_solve(t_span=[0,12],num_points=100)
By default T.plot_solution() plots the y_0, to plot general y_i use:
sage: T.plot_solution(i=0, filename=outfile)
sage: T.plot_solution(i=1, filename=outfile)
sage: T.plot_solution(i=2, filename=outfile)
The method interpolate_solution will return a spline interpolation through the points found by the solver. By default y_0 is interpolated. You can interpolate y_i through the keyword argument i.
sage: f = T.interpolate_solution()
sage: plot(f,0,12).show()
sage: f = T.interpolate_solution(i=1)
sage: plot(f,0,12).show()
sage: f = T.interpolate_solution(i=2)
sage: plot(f,0,12).show()
sage: f = T.interpolate_solution()
sage: f(pi)
0.5379...
The solver attributes may also be set up using arguments to ode_solver. The previous example can be rewritten as:
sage: T = ode_solver(g_1,y_0=[0,1,1],scale_abs=[1e-4,1e-4,1e-5],error_rel=1e-4, algorithm="rk8pd")
sage: T.ode_solve(t_span=[0,12],num_points=100)
sage: f = T.interpolate_solution()
sage: f(pi)
0.5379...
Unfortunately because python functions are used, this solver is slow on system that require many function evaluations. It is possible to pass a compiled function by deriving from the class ode_sysem and overloading c_f and c_j with C functions that specify the system. The following will work in notebook:
%cython
cimport sage.gsl.ode
import sage.gsl.ode
include 'gsl.pxi'
cdef class van_der_pol(sage.gsl.ode.ode_system):
cdef int c_f(self,double t, double *y,double *dydt):
dydt[0]=y[1]
dydt[1]=-y[0]-1000*y[1]*(y[0]*y[0]-1)
return GSL_SUCCESS
cdef int c_j(self, double t,double *y,double *dfdy,double *dfdt):
dfdy[0]=0
dfdy[1]=1.0
dfdy[2]=-2.0*1000*y[0]*y[1]-1.0
dfdy[3]=-1000*(y[0]*y[0]-1.0)
dfdt[0]=0
dfdt[1]=0
return GSL_SUCCESS
After executing the above block of code you can do the following (WARNING: the following is not automatically doctested):
sage: T = ode_solver() # not tested
sage: T.algorithm = "bsimp" # not tested
sage: vander = van_der_pol() # not tested
sage: T.function=vander # not tested
sage: T.ode_solve(y_0 = [1,0], t_span=[0,2000], num_points=1000) # not tested
sage: T.plot_solution(i=0, filename=SAGE_TMP + '/test.png') # not tested