NumPy is not imported into sage initially. To use NumPy, you first need to import it.
sage: import numpy
The basic object of computation in NumPy is an array. It is simple to create an array.
sage: l=numpy.array([1,2,3])
sage: l
array([1, 2, 3])
NumPy arrays can store any type of python object. However, for speed, numeric types are automatically converted to native hardware types (i.e., int, float, etc.) when possible. If the value or precision of a number cannot be handled by a native hardware type, then an array of Sage objects will be created. You can do calculations on these arrays, but they may be slower than using native types. When the numpy array contains Sage or python objects, then the data type is explicitly printed as object. If no data type is explicitly shown when NumPy prints the array, the type is either a hardware float or int.
sage: l=numpy.array([2**40, 3**40, 4**40])
sage: l
array([1099511627776, 12157665459056928801, 1208925819614629174706176], dtype=object)
sage: a=2.0000000000000000001
sage: a.prec() # higher precision than hardware floating point numbers
67
sage: numpy.array([a,2*a,3*a])
array([2.000000000000000000, 4.000000000000000000, 6.000000000000000000], dtype=object)
The dtype attribute of an array tells you the type of the array. For fast numerical computations, you generally want this to be some sort of float. If the data type is float, then the array is stored as an array of machine floats, which takes up much less space and which can be operated on much faster.
sage: l=numpy.array([1.0, 2.0, 3.0])
sage: l.dtype
dtype('float64')
You can create an array of a specific type by specifying the dtype parameter. If you want to make sure that you are dealing with machine floats, it is good to specify dtype=float when creating an array.
sage: l=numpy.array([1,2,3], dtype=float)
sage: l.dtype
dtype('float64')
You can access elements of a NumPy array just like any list, as well as take slices
sage: l=numpy.array(range(10),dtype=float)
sage: l[3]
3.0
sage: l[3:6]
array([ 3., 4., 5.])
You can do basic arithmetic operations
sage: l+l
array([ 0., 2., 4., 6., 8., 10., 12., 14., 16., 18.])
sage: 2.5*l
array([ 0. , 2.5, 5. , 7.5, 10. , 12.5, 15. , 17.5, 20. , 22.5])
Note that l*l will multiply the elements of l componentwise. To get a dot product, use numpy.dot().
sage: l*l
array([ 0., 1., 4., 9., 16., 25., 36., 49., 64., 81.])
sage: numpy.dot(l,l)
285.0
We can also create two dimensional arrays
sage: m = numpy.array([[1,2],[3,4]])
sage: m
array([[1, 2],
[3, 4]])
sage: m[1,1]
4
This is basically equivalent to the following
sage: m=numpy.matrix([[1,2],[3,4]])
sage: m
matrix([[1, 2],
[3, 4]])
sage: m[0,1]
2
The difference is that with numpy.array(), m is treated as just an array of data. In particular m*m will multiply componentwise, however with numpy.matrix(), m*m will do matrix multiplication. We can also do matrix vector multiplication, and matrix addition
sage: n = numpy.matrix([[1,2],[3,4]],dtype=float)
sage: v = numpy.array([[1],[2]],dtype=float)
sage: n*v
matrix([[ 5.],
[ 11.]])
sage: n+n
matrix([[ 2., 4.],
[ 6., 8.]])
If n was created with numpy.array(), then to do matrix vector multiplication, you would use numpy.dot(n,v).
All NumPy arrays have a shape attribute. This is a useful attribute to manipulate
sage: n = numpy.array(range(25),dtype=float)
sage: n
array([ 0., 1., 2., 3., 4., 5., 6., 7., 8., 9., 10.,
11., 12., 13., 14., 15., 16., 17., 18., 19., 20., 21.,
22., 23., 24.])
sage: n.shape=(5,5)
sage: n
array([[ 0., 1., 2., 3., 4.],
[ 5., 6., 7., 8., 9.],
[ 10., 11., 12., 13., 14.],
[ 15., 16., 17., 18., 19.],
[ 20., 21., 22., 23., 24.]])
This changes the one-dimensional array into a 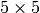 array.
array.
NumPy arrays can be sliced as well
sage: n=numpy.array(range(25),dtype=float)
sage: n.shape=(5,5)
sage: n[2:4,1:3]
array([[ 11., 12.],
[ 16., 17.]])
It is important to note that the sliced matrices are references to the original
sage: m=n[2:4,1:3]
sage: m[0,0]=100
sage: n
array([[ 0., 1., 2., 3., 4.],
[ 5., 6., 7., 8., 9.],
[ 10., 100., 12., 13., 14.],
[ 15., 16., 17., 18., 19.],
[ 20., 21., 22., 23., 24.]])
You will note that the original matrix changed. This may or may not be what you want. If you want to change the sliced matrix without changing the original you should make a copy
m=n[2:4,1:3].copy()
Some particularly useful commands are
sage: x=numpy.arange(0,2,.1,dtype=float)
sage: x
array([ 0. , 0.1, 0.2, 0.3, 0.4, 0.5, 0.6, 0.7, 0.8, 0.9, 1. ,
1.1, 1.2, 1.3, 1.4, 1.5, 1.6, 1.7, 1.8, 1.9])
You can see that numpy.arange() creates an array of floats increasing by 0.1 from 0 to 2. There is a useful command numpy.r_() that is best explained by example
sage: from numpy import r_
sage: j=numpy.complex(0,1)
sage: RealNumber=float
sage: Integer=int
sage: n=r_[0.0:5.0]
sage: n
array([ 0., 1., 2., 3., 4.])
sage: n=r_[0.0:5.0, [0.0]*5]
sage: n
array([ 0., 1., 2., 3., 4., 0., 0., 0., 0., 0.])
numpy.r_() provides a shorthand for constructing NumPy arrays efficiently. Note in the above 0.0:5.0 was shorthand for 0.0, 1.0, 2.0, 3.0, 4.0. Suppose we want to divide the interval from 0 to 5 into 10 intervals. We can do this as follows
sage: r_[0.0:5.0:11*j]
array([ 0. , 0.5, 1. , 1.5, 2. , 2.5, 3. , 3.5, 4. , 4.5, 5. ])
The notation 0.0:5.0:11*j expands to a list of 11 equally space points between 0 and 5 including both endpoints. Note that j is the NumPy imaginary number, but it has this special syntax for creating arrays. We can combine all of these techniques
sage: n=r_[0.0:5.0:11*j,int(5)*[0.0],-5.0:0.0]
sage: n
array([ 0. , 0.5, 1. , 1.5, 2. , 2.5, 3. , 3.5, 4. , 4.5, 5. ,
0. , 0. , 0. , 0. , 0. , -5. , -4. , -3. , -2. , -1. ])
Another useful command is numpy.meshgrid(), it produces meshed grids. As an
example suppose you want to evaluate 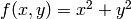 on a
an equally spaced grid with
on a
an equally spaced grid with 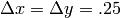 for
for
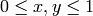 . You can do that as follows
. You can do that as follows
sage: import numpy
sage: j=numpy.complex(0,1)
sage: def f(x,y):
... return x**2+y**2
sage: from numpy import meshgrid
sage: x=numpy.r_[0.0:1.0:5*j]
sage: y=numpy.r_[0.0:1.0:5*j]
sage: xx,yy= meshgrid(x,y)
sage: xx
array([[ 0. , 0.25, 0.5 , 0.75, 1. ],
[ 0. , 0.25, 0.5 , 0.75, 1. ],
[ 0. , 0.25, 0.5 , 0.75, 1. ],
[ 0. , 0.25, 0.5 , 0.75, 1. ],
[ 0. , 0.25, 0.5 , 0.75, 1. ]])
sage: yy
array([[ 0. , 0. , 0. , 0. , 0. ],
[ 0.25, 0.25, 0.25, 0.25, 0.25],
[ 0.5 , 0.5 , 0.5 , 0.5 , 0.5 ],
[ 0.75, 0.75, 0.75, 0.75, 0.75],
[ 1. , 1. , 1. , 1. , 1. ]])
sage: f(xx,yy)
array([[ 0. , 0.0625, 0.25 , 0.5625, 1. ],
[ 0.0625, 0.125 , 0.3125, 0.625 , 1.0625],
[ 0.25 , 0.3125, 0.5 , 0.8125, 1.25 ],
[ 0.5625, 0.625 , 0.8125, 1.125 , 1.5625],
[ 1. , 1.0625, 1.25 , 1.5625, 2. ]])
You can see that numpy.meshgrid() produces a pair of matrices, here denoted
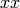 and
and 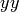 , such that
, such that ![(xx[i,j],yy[i,j])](_images/math/18caec6647b3f4d6feb38561737e2c4549984b5f.png) has coordinates
has coordinates
![(x[i],y[j])](_images/math/87f68142160153e9315770d89c27fc0913311924.png) . This is useful because to evaluate
. This is useful because to evaluate 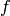 over a grid, we
only need to evaluate it on each pair of entries in
over a grid, we
only need to evaluate it on each pair of entries in 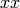 ,
, 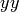 . Since
NumPy automatically performs arithmetic operations on arrays
componentwise, it is very easy to evaluate functions over a grid with
very little code.
. Since
NumPy automatically performs arithmetic operations on arrays
componentwise, it is very easy to evaluate functions over a grid with
very little code.
A useful module is the numpy.linalg module. If you want to solve an
equation 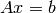 do
do
sage: import numpy
sage: from numpy import linalg
sage: A=numpy.random.randn(5,5)
sage: b=numpy.array(range(1,6))
sage: x=linalg.solve(A,b)
sage: numpy.dot(A,x)
array([ 1., 2., 3., 4., 5.])
This creates a random 5x5 matrix A, and solves 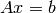 where
b=[0.0,1.0,2.0,3.0,4.0]. There are many other routines in the numpy.linalg
module that are mostly self-explanatory. For example there are
qr and lu routines for doing QR and LU decompositions. There
is also a command eigs for computing eigenvalues of a matrix. You can
always do <function name>? to get the documentation which is quite
good for these routines.
where
b=[0.0,1.0,2.0,3.0,4.0]. There are many other routines in the numpy.linalg
module that are mostly self-explanatory. For example there are
qr and lu routines for doing QR and LU decompositions. There
is also a command eigs for computing eigenvalues of a matrix. You can
always do <function name>? to get the documentation which is quite
good for these routines.
Hopefully this gives you a sense of what NumPy is like. You should explore the package as there is quite a bit more functionality.